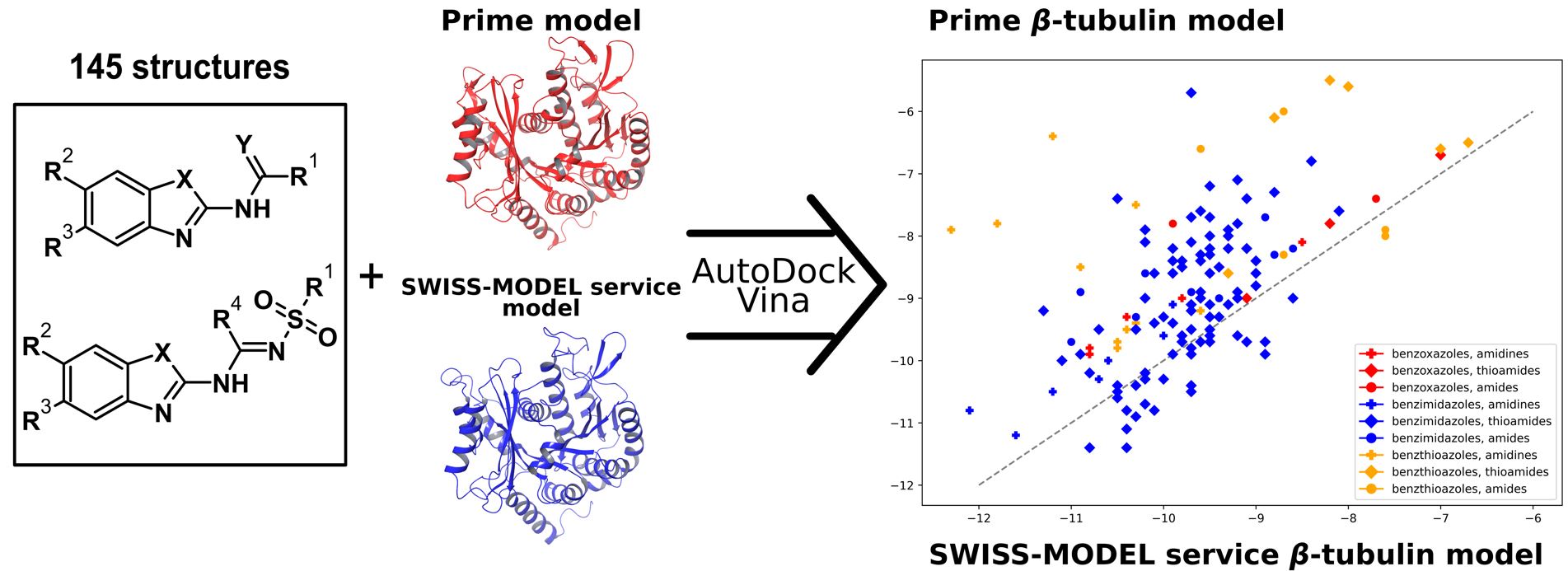
Development of A. solani β-tubulin models and comparison of docking results for benzo[d]azoles derivatives as potential antifungal agents
Abstract
Keywords
Full Text:
PDFReferences
Negi AS, Gautam Y, Alam S, Chanda D, Luqman S, Sarkar J, et al. Natural antitubulin agents: Importance of 3,4,5-trimethoxyphenyl fragment. Bioorg Med Chem. 2015;23:373–89. doi:10.1016/j.bmc.2014.12.027
Ranjan P, Kumar SP, Kari V, Jha PC. Exploration of interaction zones of β-tubulin colchicine binding domain of helminths and binding mechanism of anthelmintics. Comput Biol Chem. 2017;68:78–91. doi:10.1016/j.compbiolchem.2017.02.008
Davidse LC. Differential binding of methyl benzimidazol-2-yl carbamate to fungal tubulin as a mechanism of resistance to this antimitotic agent in mutant strains of Aspergillus nidulans. J Cell Biol. 1977;72:174–193. doi:10.1083/jcb.72.1.174
Li W, Sun H, Xu S, Zhu Z, Xu J. Tubulin inhibitors targeting the colchicine binding site: A perspective of privileged structures. Future Med Chem. 2017;9:1765–1794. doi:10.4155/fmc-2017-0100
Massarotti A, Coluccia A, Silvestri R, Sorba G, Brancale A. The Tubulin Colchicine Domain: a Molecular Modeling Perspective. ChemMedChem. 2012;7:33–42. doi:10.1002/cmdc.201100361
Wang Y, Zhang H, Gigant B, Yu Y, Wu Y, Chen X, et al. Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery. FEBS J. 2016;283:102–111. doi:10.1111/febs.13555
Tuna BG, Atalay PB, Kuku G, Acar EE, Kara HK, Yilmaz MD, et al. Enhanced antitumor activity of carbendazim on HeLa cervical cancer cells by aptamer mediated controlled release. RSC Adv. 2019;9:36005–36010. doi:10.1111/febs.13555
Goyal K, Sharma A, Arya R, Sharma R, Gupta GK, Sharma AK. Double Edge Sword Behavior of Carbendazim: A Potent Fungicide With Anticancer Therapeutic Properties. Anticancer Agents Med Chem. 2018;18:38–45. doi:10.2174/1871520616666161221113623
Yenjerla M, Cox C, Wilson L, Jordan MA. Carbendazim Inhibits Cancer Cell Proliferation by Suppressing Microtubule Dynamics. J Pharmacol Exp Ther. 2009;328:390–398. doi:10.1124/jpet.108.143537
Vela-Corcía D, Romero D, de Vicente A, Pérez-García A. Analysis of β-tubulin-carbendazim interaction reveals that binding site for MBC fungicides does not include residues involved in fungicide resistance. Sci Rep. 2018;8:7161. doi:10.1038/s41598-018-25336-5
Xu S, Wang J, Wang H, Bao Y, Li Y, Govindaraju M, et al. Molecular characterization of carbendazim resistance of Fusarium species complex that causes sugarcane pokkah boeng disease. BMC Genomics. 2019;20:115. doi:10.1186/s12864-019-5479-6
Cai M, Lin D, Chen L, Bi Y, Xiao L, Liu X. M233I Mutation in the β-Tubulin of Botrytis cinerea Confers Resistance to Zoxamide. Sci Rep. 2015;5:16881. doi:10.1038/srep16881
Aguayo-Ortiz R, Méndez-Lucio O, Medina-Franco JL, Castillo R, Yépez-Mulia L, Hernández-Luis F, et al. Towards the identification of the binding site of benzimidazoles to β-tubulin of Trichinella spiralis: Insights from computational and experimental data. J Mol Graph Model. 2013;41:12–19. doi:10.1016/j.jmgm.2013.01.007
Aguayo-Ortiz R, Méndez-Lucio O, Romo-Mancillas A, Castillo R, Yépez-Mulia L, Medina-Franco JL, et al. Molecular basis for benzimidazole resistance from a novel β-tubulin binding site model. J Mol Graph Model. 2013;45:26–37. doi:10.1016/j.jmgm.2013.07.008
Wang Y, Zhang H, Gigant B, Yu Y, Wu Y, Chen X, et al. Structures of a diverse set of colchicine binding site inhibitors in complex with tubulin provide a rationale for drug discovery. FEBS J. 2016;283:102–11. doi:10.1111/febs.13555
Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi:10.1002/jcc.21334
Schrödinger Release 2019-2: Maestro, Induced Fit Docking Protocol, MacroModel, Prime; Schrödinger, LLC: New York, NY, 2019.
Obydennov KL, Kalinina TA, Galieva NA, Beryozkina T V., Zhang Y, Fan Z, et al. Synthesis, Fungicidal Activity, and Molecular Docking of 2-Acylamino and 2-Thioacylamino Derivatives of 1H-benzo[d]imidazoles as Anti-Tubulin Agents. J Agric Food Chem. 2021;69:12048–12062. doi:10.1021/acs.jafc.1c03325
Berthold MR, Cebron N, Dill F, Gabriel TR, Kötter T, Meinl T, et al. KNIME: The Konstanz Information Miner. 2008. p. 319–326. doi:10.1007/978-3-540-78246-9_38
Sander T, Freyss J, von Korff M, Rufener C. DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. J Chem Inf Model. 2015;55:460–473. doi:10.1021/ci500588j
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi:10.1002/jcc.21256
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi:10.1186/1758-2946-3-33
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:296–303. doi:10.1093/nar/gky427
Voinkov EK, Drokin RA, Fedotov V V., Butorin II, Savateev K V., Lyapustin DN, et al. Azolo[5,1‐c][1,2,4]triazines and Azoloazapurines: Synthesis, Antimicrobial activity and in silico Studies. ChemistrySelect. 2022;7. doi:10.1002/slct.202104253
Tavella D, Ouellette DR, Garofalo R, Zhu K, Xu J, Oloo EO, et al. A novel method for in silico assessment of Methionine oxidation risk in monoclonal antibodies: Improvement over the 2-shell model. PLoS One. 2022;17:e0279689. doi:10.1371/journal.pone.0279689
Lihan M, Lupyan D, Oehme D. Target‐template relationships in protein structure prediction and their effect on the accuracy of thermostability calculations. Protein Sci. 2023;32. doi:10.1002/pro.4557
Rodriguez Moncivais OJ, Chavez SA, Estrada Jimenez VH, Sun S, Li L, Kirken RA, et al. Structural analysis of janus tyrosine kinase variants in hematological malignancies: implications for drug development and opportunities for novel therapeutic strategies. Int J Mol Sci. 2023;24:14573. doi:10.3390/ijms241914573
Champion C, Gall R, Ries B, Rieder SR, Barros EP, Riniker S. Accelerating Alchemical Free Energy Prediction Using a Multistate Method: Application to Multiple Kinases. J Chem Inf Model. 2023;63:7133–47. doi:10.1021/acs.jcim.3c01469
Berman HM. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi:10.1093/nar/28.1.235
Alhalaweh A, Lou B, Bostrom D, Velaga SP. CCDC 668711: Experimental Crystal Structure Determination. CSD Commun. 2007;668711. doi:10.5517/ccqfvb9
Lin Y, Ong YC, Keller S, Karges J, Bouchene R, Manoury E, et al. Synthesis, characterization and antiparasitic activity of organometallic derivatives of the anthelmintic drug albendazole. Dalt Trans. 2020;49:6616–6626. doi:10.1039/D0DT01107J
Hiscock JR, Gale PA, Lalaoui N, Light ME, Wells NJ. Benzimidazole-based anion receptors exhibiting selectivity for lactate over pyruvate. Org Biomol Chem. 2012;10:7780. doi:10.1039/c2ob26299a
Beisken S, Meinl T, Wiswedel B, de Figueiredo LF, Berthold M, Steinbeck C. KNIME-CDK: Workflow-driven cheminformatics. BMC Bioinformatics. 2013;14:257. doi:10.1039/c2ob26299a
National Center for Biotechnology Information (NCBI)[Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. 1988. https://www.ncbi.nlm.nih.gov/
Gui M, Croft JT, Zabeo D, Acharya V, Kollman JM, Burgoyne T, et al. SPACA9 is a lumenal protein of human ciliary singlet and doublet microtubules. Proc Natl Acad Sci. 2022;119. doi:10.1073/pnas.2207605119
DOI: https://doi.org/10.15826/chimtech.2024.11.1.04
Copyright (c) 2023 Konstantin L. Obydennov, Tatiana A. Kalinina, Tatiana V. Glukhareva, Vasiliy A. Bakulev

This work is licensed under a Creative Commons Attribution 4.0 International License.
Chimica Techno Acta, 2014–2025
eISSN 2411-1414
Copyright Notice







